| SBP |
|---|
What is a SBP?
A Synthetic Binding Protein (SBP) is a human-made protein binder that has not been found in nature and been tailored to bind to molecular target of interest. A kind of SBPs with similar characteristics are usually developed based on the same parent protein (naturally occurring or synthetic) or protein scaffold.
When given a certain target, SBPs can be generated through protein engineering and design technologies through introducing altered amino acids or sequence insertions on the selected parent protein/protein scaffold (Figure 1).

Figure 1. Flowchart of the synthetic binding proteins (SBPs) generation through protein engineering and design.
Key properties of SBPs
There are two types of SBPs in SYNBIP: non-antibody and antibody fragment. Compared with small molecules and classical antibodies, SBPs offer the greater stability, smaller size, less immunogenic, better tissue penetration, and so on (Figure 2).
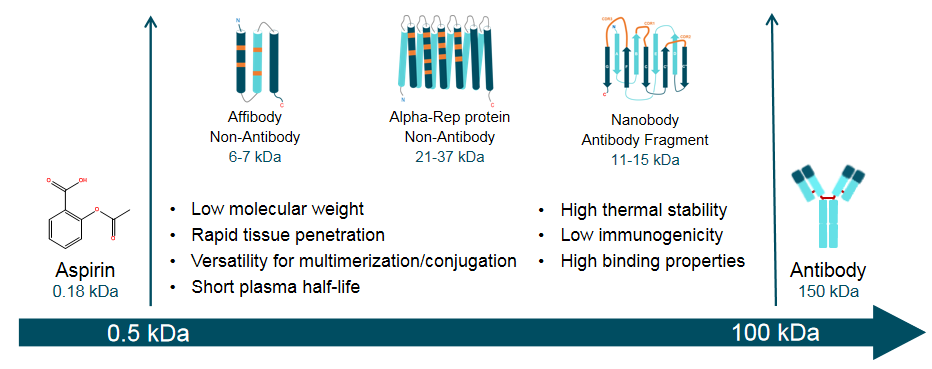
Figure 2. Key properties of the synthetic binding proteins (SBPs) compared with small molecules and classical antibodies.
| Latest Release |
|---|
SYNBIP Database 2.0 contains 68 protein scaffolds, 2,264 unique SBPs, 899 SBP epitopes, 12,205 sequences in expanding SBP space, 78,473 and 16,401,555 protein domains with SBP-like scaffolds from the RCSB PDB and AlphaFold DB. 62.4% of SBPs are shorter than 300 amino acids, encompassing all types of protein folds, including α-helices, β-sheets, and loops. The sources of SBP include peer-reviewed literature, clinical databases, and listed approved SBPs published on pharmaceutical company websites.The binding targets of all SBPs include 555 protein targets, 28 small molecular targets, and 26 other targets (such as carbohydrates, RNAs, DNAs, etc.). For protein scaffolds, the numbers of non-antibody scaffolds and antibody fragments are 57 and 11, respectively. For SBPs, 2,016, 209, and 39 SBPs collected are identified with 1, 2, and ≥3 binding targets, respectively. In addition, 1,751 SBPs (~67.2%) are with binding affinities reported.
| SYNBIP 2.0 |
|---|
A major update of SYNBIP was therefore performed in this work. First, the epitopes of 870 representative SBPs from each scaffold were structurally mapped by identifying structures of corresponding SBPs in complex with their protein targets. Those structures come from experimentally determined coordinates stored in RCSB PDB or computationally predicted models constructed using well-established protocol.Second, to expand the sequence space of SBPs, 11,025 new synthetic proteins covering 55 different scaffolds were designed through DL based framework ProteinMPNN based on SBP monomers and SBP-target complexes collected in SYNBIP and with their physicochemical properties analyzed comprehensively. Third, 78,473 and 16,401,555 protein domains with SBP-like scaffolds were identified by Foldseek from the RCSB PDB and AlphaFold DB, respectively. The identified proteins come from 54,736 different organisms. Finally, routine updates were performed on this database, with over 500 new SBPs added, covering 65% (44/68) of the scaffolds, and all SBP structures were updated using AlphaFold2, along with updates on their clinical status.
| SYNBIP |
|---|
Synthetic Binding Protein Database (SYNBIP) is a comprehensive, free-to-access, online database, which comprehensively describes thousands of SBPs from the perspectives of scaffolds, biophysical and functional properties. Detailed data are provided, including the targets, sequences, structures, and application information of SBPs. Currently, SBPs have been applied in three main areas: research (e.g. tools for stabilizing protein structures) diagnosis (e.g. imaging agents) and, therapeutics (e.g. COVID-19 and cancers).
Sequence and Structure Information of SBPs
There are 1359 SBPs in SYNBIP with full-length sequence information, which account for most (64.4%) of those 2,111 collected SBPs. The collected structures of 246 SBPs were resolved by the technique of nuclear magnetic resonance (NMR), X-ray crystallography (X-Ray), or cryogenic electron microscopy (Cryo-EM). In addition, the 3D structures of 1083 SBPs without any experimentally validated structure were modelled using trRosetta, and the confidence estimation scores (TM-scores) of all predicted SBP structures are higher than 0.7, indicating a correctly modelled topology.
| Please Enter your keyword |
|---|
In the field of “Please enter your keyword and search for SBP”, users can find Synthetic Binding Protein (SBP) entries by searching SBP name or synonym, protein scaffold name and target name among the entire textual component of SYNBIP. If you want Protein Scaffold (PSS) entries or Binding Target (BTS) entries, you can also search scaffold name, SBP name or target name and select the corresponding button below the textual component.
Query can be submitted by entering keywords into the main searching frame. Users can specify part or full name in the text field. To facilitate a more customized input query, the wildcard characters of “*”, “-” and “?” are also supported in SYNBIP.
(1) If search “Monobody BMS-962476” and click the “Synthetic Binding Protein (SBP)” button, find two entries with SBP name “Monobody BMS-962476” and “Monobody BMS-986192”. If you want only entry with SBP name “Monobody BMS-962476”, double quotation marks are required when you search the SBP name;
(2) If search “Monobody” and click the “Protein Scaffold (PSS)” button, find only entry with PSS name “Monobody”;
(3) If search “EGFR” and click the “Binding Target (BTS)” button, find related entries with BTS name “Epidermal growth factor receptor” or “EGFR”;
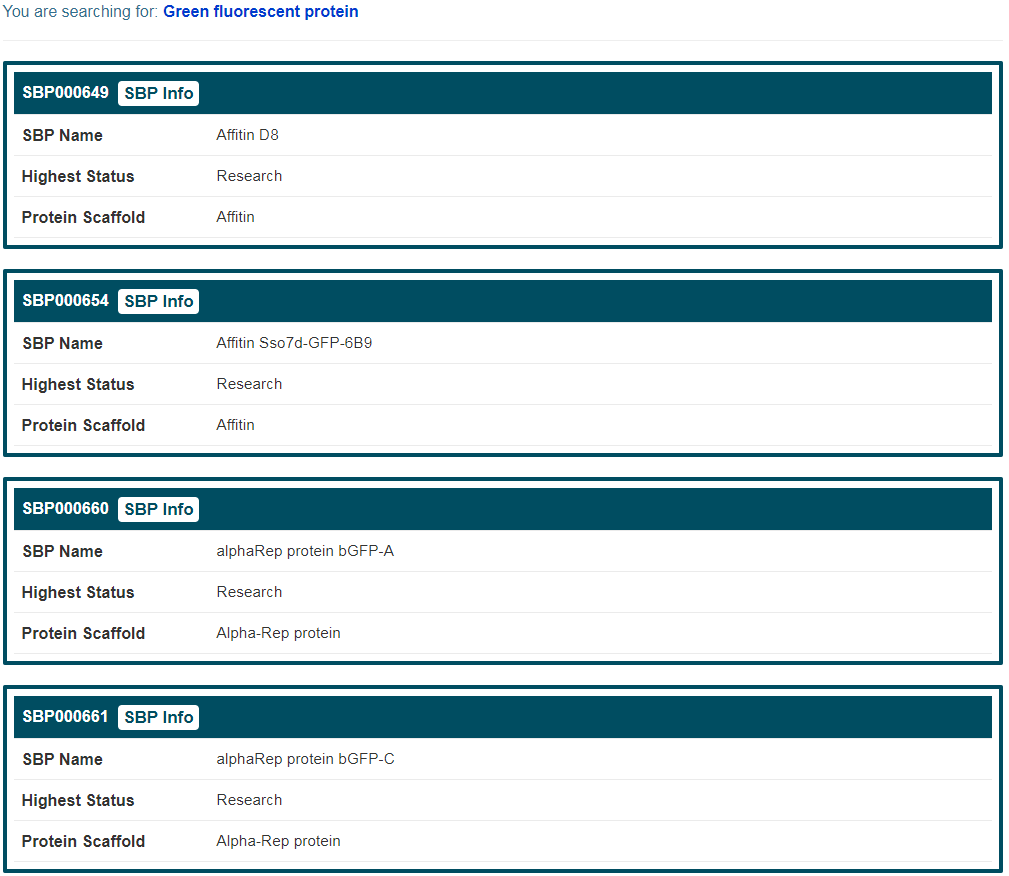
(4) If search “Green fluorescent protein”, a target name and click the “Synthetic Binding Protein (SBP)” button, find entries with the SBP names including “Affitin anti-GFP D8”, “alphaRep protein anti-GFP clone A”, and so on, all of which bind to the target “Green fluorescent protein”;
(5) If search “BMS-962476” and click the SBP button, four entries “Monobody BMS-962476”, “Monobody BMS-986192”, “Monobody Pegdinetanib” and “Monobody Talditercept alfa” will be find. Synonyms of the latter two SBPs contain “BMS”. Here “-” is equal to the word “or”;
(6) If search “Fynomer anti-Chymase 4C-?4” and click the SBP button, find entries with SBP names like “Fynomer anti-Chymase 4C-A4” and “Fynomer anti-Chymase 4C-E4”. Here “?” represents any single English letter and is only effective in a phrase consisting of two or more words;
(7) If search “BMS*” and click the SBP button, find all SBPs whose SBP name or synonyms have this word “BMS”. Here “*” represents an arbitrary string.
| Search for Synthetic Binding Protein (SBP) |
|---|
In the field of “Search for Synthetic Binding Protein (SBP)”, users can find SBP entries by selecting SBP name, protein scaffold name and target name.
1. Search for SBP by SBP Name
In this field of “Search for SBP by SBP Name”, users can select one of SBP names and get corresponding SBP information.
For example: if search “Monobody BMS-962476”, users can access to basic information of the SBP. If you want to know more about this SBP, you can click the corresponding “SBP Info” button.


2. Search for SBP by Protein Scaffold Name
In this field of "Search for SBP by Protein Scaffold Name", users can select one of protein scaffolds and get corresponding SBP information of this scaffold.
For example: if search "Monobody", users can access to corresponding SBP entries of the scaffold. If you want to know more about SBP, you can click the corresponding "SBP Info" button.

3. Search for SBP by Target Name
In this field of "Search for SBP by Target Name", users can select one of targets and get corresponding SBP information of this target.
For example: if select "Tumor necrosis factor", users can access to corresponding SBP entries of the target. If you want to know more about this SBP, you can click the corresponding "SBP Info" button.

4. SBP information
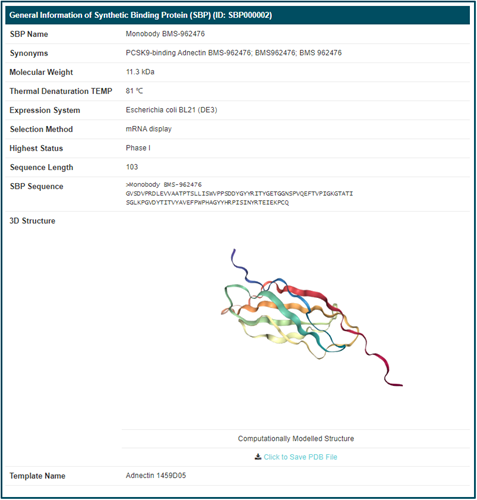
By clicking the “SBP Info” button, the detailed information page of “Monobody BMS-962476” will be displayed. Details of SBP are divided into four parts, “General Information of SBP”, “Protein Scaffold Information of This SBP”, “Binding Target(s) of This SBP” and “Clinical Trial Information of This SBP”. The general information of “Monobody BMS-962476” includes its “SBP Name”, “Synonyms”, “Molecular Weight”, “Thermal Denaturation TEMP”, "Expression System", “Selection Method”, “Highest Status”, “PDB ID”, “Sequence Length”, “SBP Sequence”, “3D Structure” (Computationally modelled structure or Experimentally validated structure), “Template Name” and "Template Sequence". All computational modeling structures are obtained from AlphaFold2. Additionally, you can download and save PDB file of this SBP.
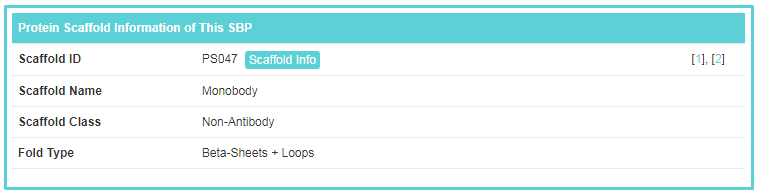
“Protein Scaffold Information of This SBP” includes general information of the protein scaffold corresponding to this SBP. The scaffold information is important to SBP, because a protein scaffold represents a kind of SBPs with similar characteristics and embodies fundamental law of the SBP. If you want to know more about this scaffold, you can click the “Scaffold Info” button.
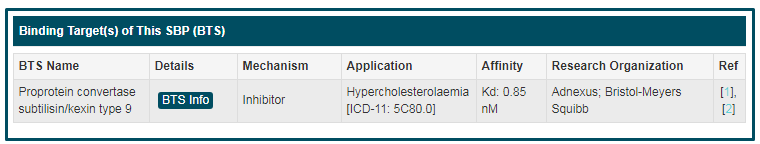
“Binding Target(s) of This SBP (BTS)” includes relational information between the target and this SBP, including includes “Target Name”, “Details”, “Mechanism”, “Application”, “Affinity” and “Research Organization”. “Mechanism” means the action between targets and this SBP. Among the values of “mechanism”, “binder” represents the binding of SBP to the target in order to promote crystallization, recognize molecules, and so on. If you want to know more about this target, you can click the “BTS Info” button.
“Clinical Trial Information of This SBP” includes “Registration Number”, “Title”, “Indication”, “Phase”, “Status” and “Sponsor”. If you want to know more about clinical information of this SBP, you can link to other related databases by clicking the “Registration Number”.

| Search for Protein Scaffold of SBP (PSS) |
|---|
In the field of "Search for PSS", users can find PSS entries by selecting PSS name, SBP name and parental protein name.
1. Search for PSS by PSS Name
In this field of "Search for PSS by PSS Name", users can select one of protein scaffolds and get the PSS information.
For example: if search “Affibody”, users can access to basic information of the PSS. If you want to know more about this PSS, you can click the corresponding "PSS Info" button.


2. Search for PSS by SBP Name
In this field of "Search for PSS by SBP Name", users can select one of SBPs and get corresponding PSS information of this SBP.
For example: if select “Monobody BMS-962476”, users can access to corresponding PSS entry of the SBP. If you want to know more about this PSS, you can click the corresponding "PSS Info" button.


3. Search for PSS by Parental Protein Name
In this field of "Search for PSS by Parental Protein Name", users can select one of parental proteins and get corresponding PSS information.
For example: if select "10th domain of Fibronectin III", users can access to corresponding PSS entry of the parental protein. If you want to know more about this PSS, you can click the corresponding "PSS Info" button.


| 4. PSS information |
|---|
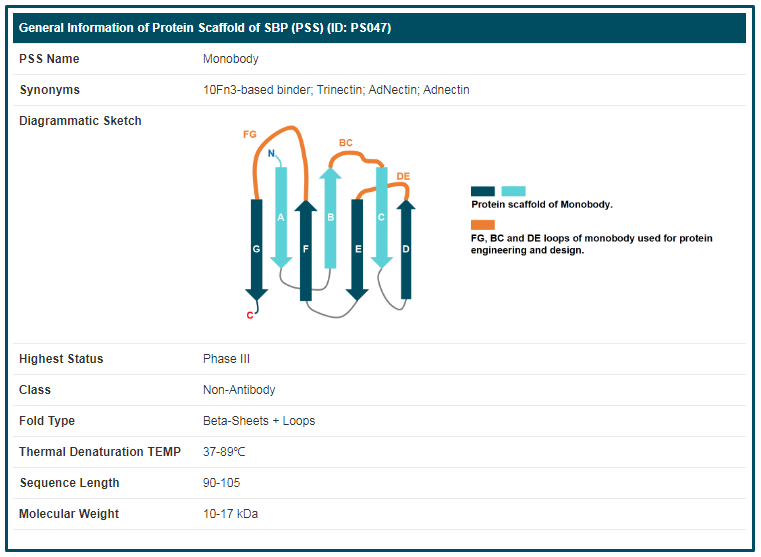
By clicking the “PSS Info” button, the detailed information page of “Monobody” will be displayed. Details of PSS are divided into three parts, “General Information of PSS”, “Parental Protein of This PSS” and “Synthetic Binding Protein (SBP) Featuring this PSS”. The general information of “Monobody” includes its “PSS Name”, “Synonyms”, “Diagrammatic Sketch”, “Highest Status”, "Class", “Fold Type”, “Thermal Denaturation TEMP”, “Sequence Length” and “Molecular Weight”.
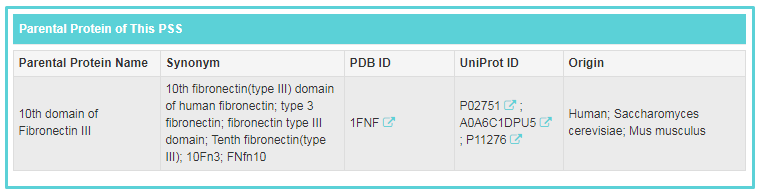
“Parental Protein of This PSS” includes “Parental Protein Name”, “Synonym”, “PDB ID”, “UniProt ID” and “Origin”. If you want to know more about this parental protein, you can link to other related databases by clicking the IDs.
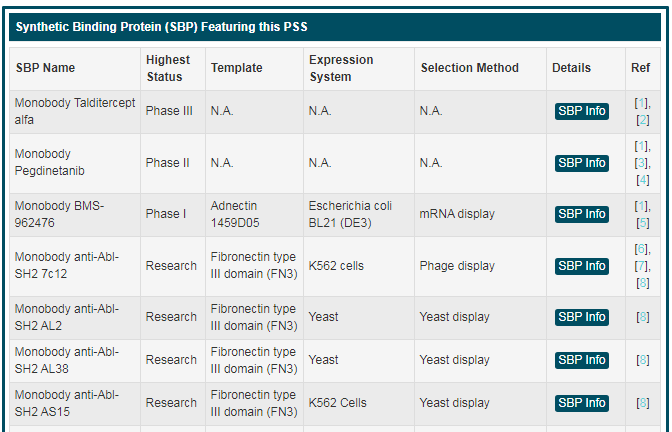
“Synthetic Binding Protein (SBP) Featuring this PSS” includes all SBPs belonging to this PSS. Here, a statistical table is displayed. Users can get information of “SBP Name”, “Highest Status”, “Template”, “Expression System” and “Selection Method”. If you want to know more about a SBP, you can click the “SBP Info” button.
| Search for Binding Target of SBP (BTS) |
|---|
In the field of "Search for Binding Target of SBP (BTS)", users can find BTS entries by selecting BTS name, BTS family name and SBP name.
1. Search for BTS by BTS Name
In this field of “Search for BTS by BTS Name”, users can select one of binding targets and get the BTS information.
For example: if search “Epidermal growth factor receptor”, users can access to basic information of the BTS from different organisms. If you want to know more about the BTS, you can click the corresponding “BTS Info” button.

2. Search for BTS by BTS Family Name
In this field of “Search for BTS by BTS Family Name”, users can select one of BTS families and get corresponding BTS information of the target family.
For example: If select “Tumor necrosis factor family”, users can access to corresponding BTS entries of the family. If you want to know more about a BTS, you can click the corresponding "BTS Info" button.

3. Search for BTS by SBP Name
In this field of "Search for BTS by SBP Name", users can select one of SBPs and get corresponding BTS information.
For example: if select “Monobody BMS-962476”, users can access to corresponding BTS entries of the SBP. If you want to know more about this BTS, you can click the corresponding “BTS Info” button.

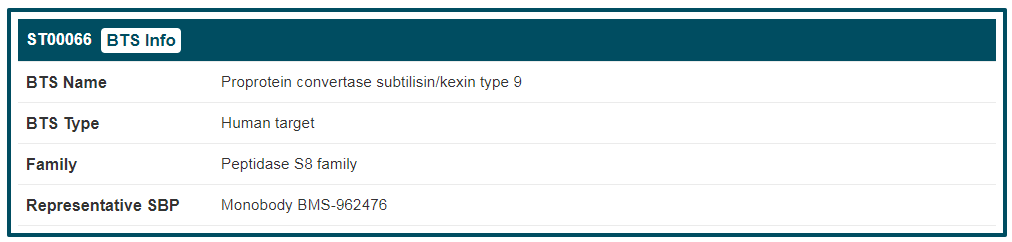
4. BTS information
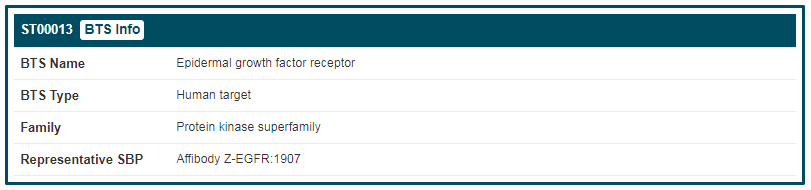
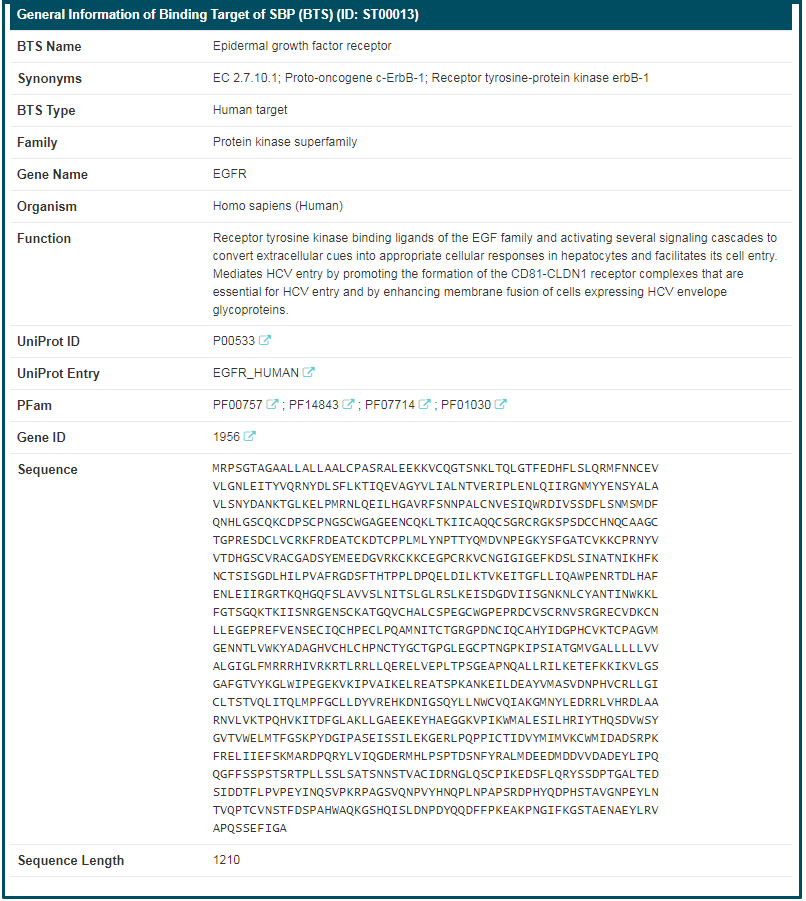
By clicking the “BTS Info” button, the detailed information page of “Epidermal growth factor receptor” will be displayed. Details of BTS are divided into two parts, “General Information of BTS” and “Synthetic Binding Protein (SBP) Targeting This BTS”. The general information of “Epidermal growth factor receptor” includes its “BTS Name”, “Synonyms”, “BTS Type”, “Family”, “Gene Name”, “Organism”, “Function”, “UniProt ID”, “UniProt Entry”, “PFam”, “Gene ID”, “Sequence” and “Sequence Length”. If you want to know more about this target, you can link to other related databases by clicking the corresponding IDs.
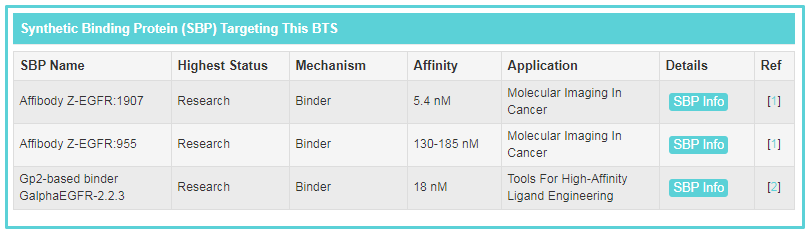
“Synthetic Binding Protein (SBP) Targeting This BTS” includes all of SBPs binding to this target. Here, a statistical table is displayed. Users can get information of “SBP Name”, “Highest Status of SBP”, “Mechanism of SBP to the target”, “Affinity between SBP and the target” and “Application of SBP”. If you want to know more about a SBP, you can click the “SBP Info” button.
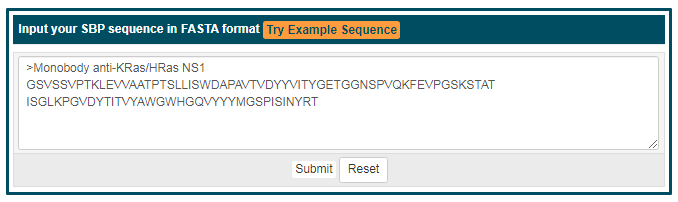
| SBP Sequence Similarity Search |
|---|
Besides traditional keywords search, we also supply SBP sequence similarity query for searching similar sequences of SBPs in SYNBIP. The similarity degree of identified SBPs will be evaluated by BLAST program, and with the sequence identities listed out. Using “Monobody anti-KRas/HRas NS1” as an example, 226 SBP sequences will be identified with identities from 28% to 100% (itself). Links to the detail information of identified targets are also provided.

| SBP Binding Target Sequence Similarity Search |
|---|
Besides traditional keywords search, we also supply SBP target sequence similarity query for searching similar sequences against all SBP targets with available sequence information in SYNBIP. The similarity degree of those identified SBP targets will be evaluated by BLAST program, and with the sequence identities listed out. Using “Human GTPase HRas” as an example, 2 target sequences will be identified with identities of 87% and 100% (itself), respectively. Links to the detail information of identified targets are also provided. By using the found targets of interested, users can select suitable SBPs for protein function and function study.

| Search the SBPs for COVID-related Studies |
|---|
In the field, users can find SBP entries applying to COVID-19 infection by searching a SBP or a protein scaffold which relate to the coronavirus.

1. Search for SBP by SBP Name
In this field of “Search for SBP by SBP Name”, users can select one of SBPs which relate to COVID-19 and get the detail SBP information.
For example: if search “Human VH dAb anti-SARS-CoV-2 B01”, users can access to basic information of the SBP and find that the application of this SBP is “SARS-CoV-2 infection (COVID-19)”.

2. Search for SBP by Protein Scaffold Name
In this field of “Search for SBP by Protein Scaffold Name”, users can select one of protein scaffolds which relate to COVID-19 and get the detail SBP information.
For example: if select “Monobody”, users can access to corresponding SBP entries which relate to COVID-19 and find that the application of all of the SBPs are “SARS-CoV-2 infection (COVID-19)”.

| Search for SBP-Target Complex |
|---|
In the field, users can find SBP-Target complex entries which display SBP Epitopes.

1. Search for SBP-Target Complex by Complex Name
In this field of “Search for SBP-Target Complex by Complex Name”, users can select one of SBP-Target complex and get the detail complex information.
For example: if search “ABD-derived affinity protein anti-ERBB2/HSA clone 1 with HSA”, users can access to basic information of the complex.
2. Search for SBP-Target Complex by SBP Name
In this field of “Search for SBP-Target Complex by SBP Name”, users can select one of SBPs and get the corresponding complex information.
For example: if search “ABD-derived affinity protein anti-ERBB2/HSA clone FACS-10”, users can access to corresponding information of the complex.

3. Search for SBP-Target Complex by Protein Scaffold Name
In this field of “Search for SBP-Target Complex by Protein Scaffold Name”, users can select one of protein scaffolds and get the corresponding complexes information.
For example: if search “ABD-derived affinity protein”, users can access to corresponding information of the complexes.

4. Search for SBP-Target Complex by Target Name
In this field of “Search for SBP-Target Complex by Target Name”, users can select one of targets and get the corresponding complexes information.
For example: if search “Adhesion G-protein coupled receptor G1”, users can access to corresponding information of the complexes.

5. SBP-Target Complex information
By clicking the “SBP-Target Complex Info” button, the detailed information page of “Affibody anti-CTX-M-15 A5888 with Beta-lactamase” will be displayed. The “Synthetic Binding Protein (SBP)-Target Complex” includes its “Complex Name”, “SBP”, “Target”, “3D structure”, “Statistics of The Docking Results”, “Complex Method”, “Complex Quality”, “SBP Structure Type” and “Target Structure Type”. If you want to know more about corresponding SBP or target, you can link to detailed page by clicking the corresponding name. Additionally, you can download and save PDB and PSE file of this SBP-Target Complex. The PSE file contains interactions predicted by PLIP. The cyan dots in the docking statistics chart represent the corresponding RosettaDock scores of the displayed structure.
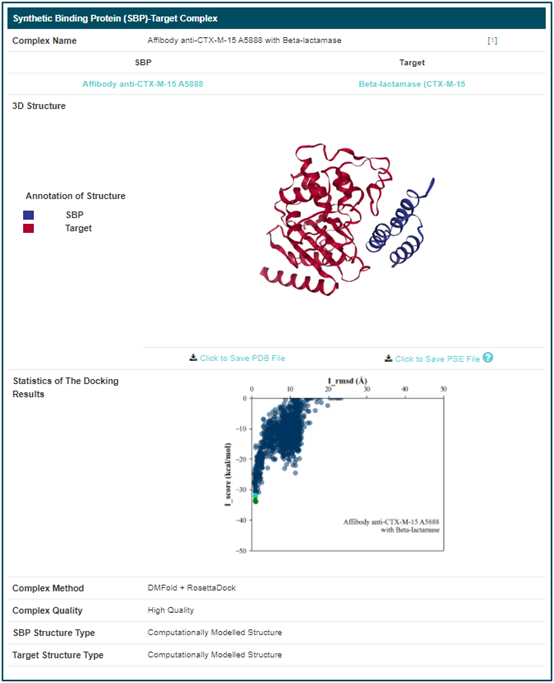
Here, the sequences designed based on this composite is also provided.
| Search the Sequences in Expanding SBP Space |
|---|
In the field, users can find sequences designed based on SBP and some parameters.

1.Search for Expanding SBP Sequence Space by SBP Name
In this field of “Search for Expanding SBP Sequence Space by SBP Name”, users can select one of SBPs and get the detail expanding sequences information.
For example: if search “ABD-derived affinity protein anti-ERBB2/HSA clone 1”, users can access to basic information of the expanding sequences.

2.Search for Expanding SBP Sequence Space by Protein Scaffold Name
In this field of “Search for Expanding SBP Sequence Space by Protein Scaffold Name”, users can select one of protein scaffolds and get the detail expanding sequences information.
For example: if search “VL dAb”, users can access to basic information of the expanding sequences”.

3.Search for Expanding SBP Sequence Space by Potential Target Name
In this field of “Search for Expanding SBP Sequence Space by Potential Target Name”, users can select one of targets and get the detail expanding sequences information.
For example: if search “CD276 antigen”, users can access to basic information of the expanding sequences.


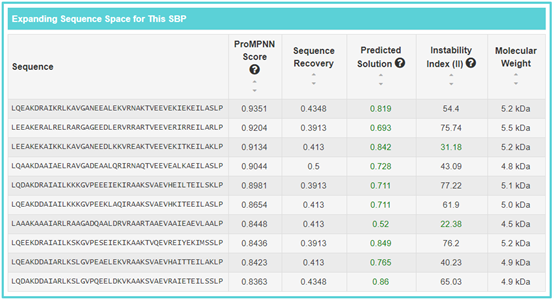
4. Expanding SBP Sequences information
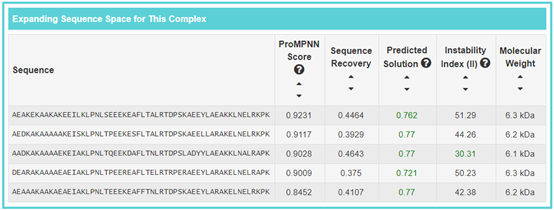
By clicking the “SBP Sequence Space Info” button, the detailed information page of “ABD-derived affinity protein anti-ERBB3/HSA clone 1” will be displayed. “Expanding Sequence Space for This SBP” refers to the designed sequence information form the SBP, including “Sequence”, “ProMPNN Score”, “Sequence Recovery”, “Predicted Solution”, “Instability Index (II)” and “Molecular Weight”. If you want to sort numerical values, you can click the corresponding section.
Sequence recovery: an important criterion for de novo protein design evaluation.
ProMPNN Score: abbreviation of “ProteinMPNN score”. It’s refers to average over residues that were designed negative log probability of sampled amino acids (Lower score is better, the score represents model's uncertainty about the predictions).
Predicted Solution: the predicted solubility value of Protein-Sol. Predicted solution value greater than 0.45 is predicted to have a higher solubility than the average soluble E.coli protein from the experimental solubility dataset, and any protein with a lower scaled solubility value is predicted to be less soluble. The predicted solubility value greater than 0.45 has been marked in green.Instability Index (II): an estimate of the stability of the sequenced in a test tube. Instability index (II) is smaller than 40 is predicted as stable, a value above 40 predicts that the protein may be unstable. The Instability Index (II) value less than 40 has been marked in green.
| Search SBP-like Protein Scaffolds |
|---|
In the field, users can find SBP-like Protein Scaffolds of one protein scaffold.

1. Search for PSS by PSS Name
In this field of “Search for PSS by PSS Name”, users can find one SBP scaffold with various SBP-like Protein Scaffolds by searching PSS name.
For example: if search “ABD-derived affinity protein”, users can access to basic information of the PSS. Then you can click the corresponding “PSS Info” button to get the information of SBP-like Protein Scaffolds.

2.Search for PSS by SBP Name
In this field of “Search for PSS by SBP Name”, users can find one SBP scaffold with various SBP-like Protein Scaffolds by searching SBP name.
For example: if search “ABD-derived affinity protein anti-ERBB2/HSA clone FACS-17”, users can access to basic information of the PSS of this SBP. Then you can click the corresponding “PSS Info” button to get the information of SBP-like Protein Scaffolds.

3.Search for PSS by Parental Protein Name
In this field of “Search for PSS by SBP Name”, users can find one SBP scaffold with various SBP-like Protein Scaffolds by searching Parental Protein name.
For example: if search “10 kDa heat shock protein, mitochondrial”, users can access to basic information of the PSS of this Parental Protein. Then you can click the corresponding “PSS Info” button to get the information of SBP-like Protein Scaffolds.

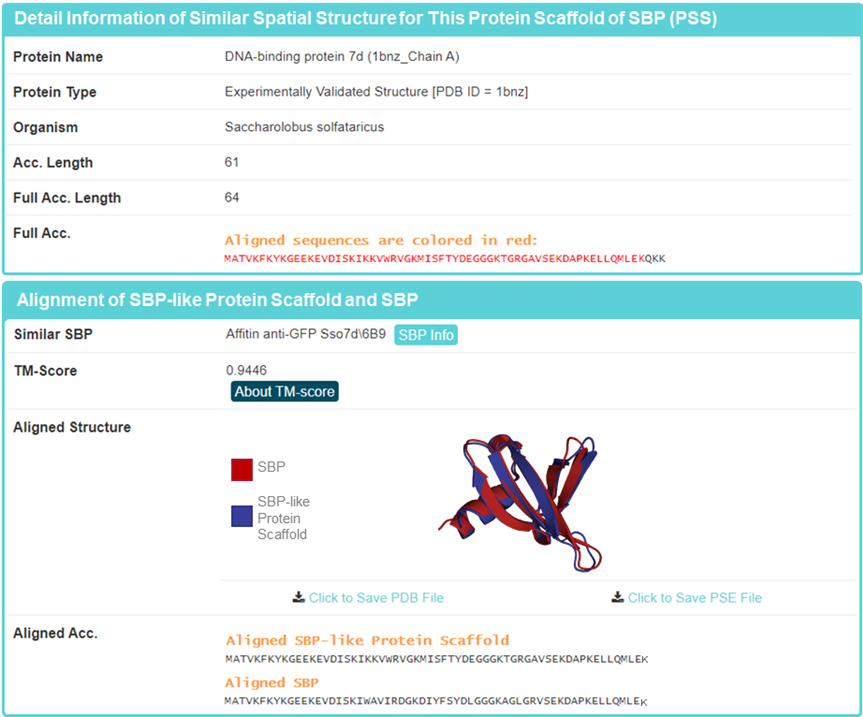
4. SBP-like protein scaffolds information
Displayed information on representative SBP-like protein scaffolds, as well as downloadable full SBP-like protein scaffolds.
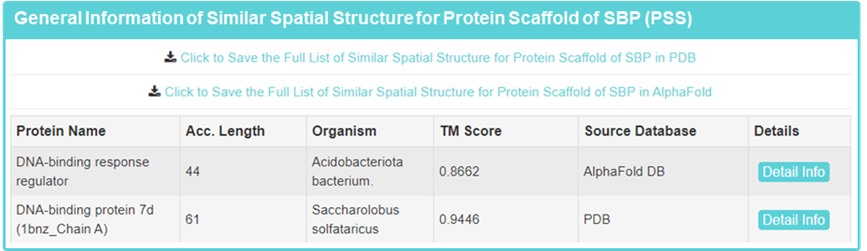
Detailed presentation of representative SBP-like protein scaffold, including “Protein Name”, “Protein Type”, “Organism”, “Acc.Length”, “Full Acc.Length “, “Full Acc.” Similar SBP”, “TM-Score”, Aligned Structure”, and “Aligned Acc.”
Acc.Length: Length of the protein fragment from the first to the last aligned amino acid. Full Acc.Length: Full-length of SBP-like protein scaffold.
TM-score: TM-score value scales the structural similarity; The larger the value, the higher the similarity is.”
Aligned Acc.: Aligned sequence information of SBP-like protein scaffold and SBP.