| SBP |
|---|
What is a SBP?
A Synthetic Binding Protein (SBP) is a human-made protein binder that has not been found in nature and been tailored to bind to molecular target of interest. A kind of SBPs with similar characteristics are usually developed based on the same parent protein (naturally occurring or synthetic) or protein scaffold.
When given a certain target, SBPs can be generated through protein engineering and design technologies through introducing altered amino acids or sequence insertions on the selected parent protein/protein scaffold (Figure 1).

Figure 1. Flowchart of the synthetic binding proteins (SBPs) generation through protein engineering and design.
Key properties of SBPs
There are two types of SBPs in SYNBIP: non-antibody and antibody fragment. Compared with small molecules and classical antibodies, SBPs offer the greater stability, smaller size, less immunogenic, better tissue penetration, and so on (Figure 2).
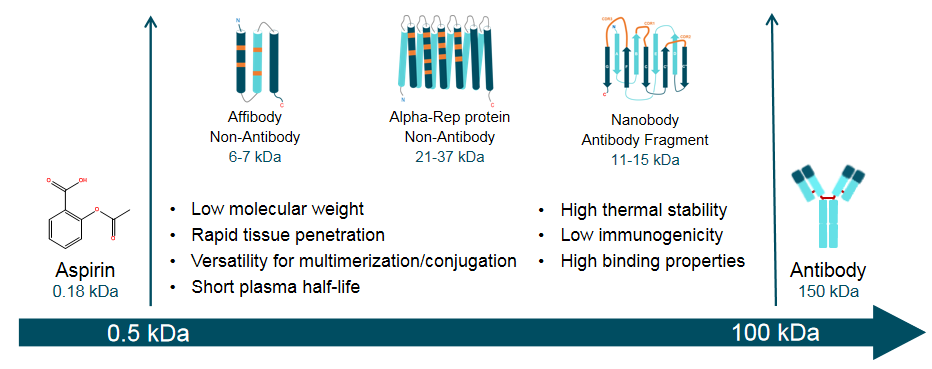
Figure 2. Key properties of the synthetic binding proteins (SBPs) compared with small molecules and classical antibodies.
| SYNBIP |
|---|
Synthetic Binding Protein Database (SYNBIP) is a comprehensive, free-to-access, online database, which comprehensively describes thousands of SBPs from the perspectives of scaffolds, biophysical and functional properties. Detailed data are provided, including the targets, sequences, structures, and application information of SBPs. Currently, SBPs have been applied in three main areas: research (e.g. tools for stabilizing protein structures) diagnosis (e.g. imaging agents) and, therapeutics (e.g. COVID-19 and cancers).
Sequence and Structure Information of SBPs
There are 1359 SBPs in SYNBIP with full-length sequence information, which account for most (65.5%) of those 2,074 collected SBPs. The collected structures of 246 SBPs were resolved by the technique of nuclear magnetic resonance (NMR), X-ray crystallography (X-Ray), or cryogenic electron microscopy (Cryo-EM). In addition, the 3D structures of 1083 SBPs without any experimentally validated structure were modelled using trRosetta, and the confidence estimation scores (TM-scores) of all predicted SBP structures are higher than 0.7, indicating a correctly modelled topology.
| SYNBIP 2.0 |
|---|
A major update of SYNBIP was therefore performed in this work. First, the epitopes of 306 representative SBPs from each scaffold were structurally mapped by identifying structures of corresponding SBPs in complex with their protein targets. Those structures come from experimentally determined coordinates stored in RCSB PDB51 or computationally predicted models constructed using well-established protocol52. Second, to expand the sequence space of SBPs, 7,245 new synthetic proteins covering 55 different scaffolds were designed through DL based framework ProteinMPNN based on the 1,327 original SBPs collected in SYNBIP4 and with their physicochemical properties analyzed comprehensively. Third, 315 of SBPs belonging to 22 new and 37 known protein scaffolds that developed in recent two years were collected by literature review in PubMed. Therefore, the systematical data refers to this majority update of SYNBIP laid a solid foundation for the rational protein engineering & design to discover of novel functional binders in the future.
| Latest Release |
|---|
SYNBIP Database 2.0 contains 68 protein scaffolds and 2074 unique SBPs. The binding targets of all SBPs include 423 protein targets, 28 small molecular targets, and 15 other targets (such as carbohydrates, RNAs, DNAs, etc.). For protein scaffolds, the numbers of non-antibody scaffolds and antibody fragments are 57 and 11, respectively. For SBPs, 1860, 192, and 22 SBPs collected are identified with 1, 2, and ≥3 binding targets, respectively. In addition, 1384 SBPs (~66.7%) are with binding affinities reported.